CRISPR
So, CRISPR, what of it? The best way to understand it is to look at the bacterial mechanism from which it originates.
We often think of bacteria as sneaky little buggers that are hell-bent on invading our bodies and giving us some nasty infection. We forget sometimes that they are living entities engaged in their own fight for survival, attacked on all sides from our body’s defences, chemical attack from disinfectants and other bacteria, being eaten by bigger bacteria – makes you feel quite sorry for them doesn’t it? They are also attacked by viruses and this is where it gets interesting – honestly. A virus is a really simple beasty, consisting essentially of a bit of DNA (or RNA) and a sugar coating which is mostly just a mechanism for getting that DNA into a living cell. No doubt this is way over-simplified but bear with me. The virus doesn’t have a mechanism for replicating its DNA so it injects it into a foreign, living cell and induces that cell to replicate it – over and over again, ultimately leading to the host cell bursting and releasing multiple copies of the virus which then infect other cells which burst releasing .... No wonder you feel ill when you catch a cold (virus)!
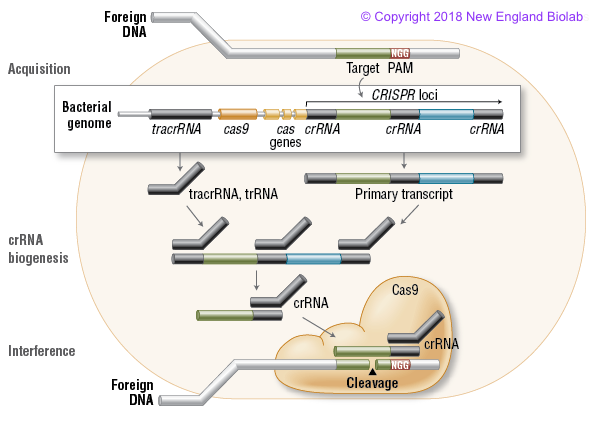
Obviously, any kind of mechanism that helps protect the bacteria from a virus must help the virus and make it more likely to thrive (natural selection etc.). This is where the CRISPR/cas mechanism comes in. It was discovered that within the DNA of certain bacterias, there occurred repeated palindromic sequences (hence the name). These sequences, each of around 20 to 40 base pairs long, are separated by short DNA (or RNA equivalent) 'spacers', each of around 20 to 70 base pairs. The spacer DNA turned out to be, in effect, bits of DNA from viruses that had previously attacked the bacteria - or its ancestors - and, it appears, provide a way for the cell to identify an hostile injection of similar viral DNA. In other words, the CRISPR sequences could be part of a bacterial immunity process. The mechanism completed by nearby 'cas' genes (CRISPR Associated Systems).
There are a number of slightly different mechanisms but the one that seems to be most studied and used for gene editing is cas9. My highly simplified understanding of how the defence works goes something like this:
We often think of bacteria as sneaky little buggers that are hell-bent on invading our bodies and giving us some nasty infection. We forget sometimes that they are living entities engaged in their own fight for survival, attacked on all sides from our body’s defences, chemical attack from disinfectants and other bacteria, being eaten by bigger bacteria – makes you feel quite sorry for them doesn’t it? They are also attacked by viruses and this is where it gets interesting – honestly. A virus is a really simple beasty, consisting essentially of a bit of DNA (or RNA) and a sugar coating which is mostly just a mechanism for getting that DNA into a living cell. No doubt this is way over-simplified but bear with me. The virus doesn’t have a mechanism for replicating its DNA so it injects it into a foreign, living cell and induces that cell to replicate it – over and over again, ultimately leading to the host cell bursting and releasing multiple copies of the virus which then infect other cells which burst releasing .... No wonder you feel ill when you catch a cold (virus)!
Obviously, any kind of mechanism that helps protect the bacteria from a virus must help the virus and make it more likely to thrive (natural selection etc.). This is where the CRISPR/cas mechanism comes in. It was discovered that within the DNA of certain bacterias, there occurred repeated palindromic sequences (hence the name). These sequences, each of around 20 to 40 base pairs long, are separated by short DNA (or RNA equivalent) 'spacers', each of around 20 to 70 base pairs. The spacer DNA turned out to be, in effect, bits of DNA from viruses that had previously attacked the bacteria - or its ancestors - and, it appears, provide a way for the cell to identify an hostile injection of similar viral DNA. In other words, the CRISPR sequences could be part of a bacterial immunity process. The mechanism completed by nearby 'cas' genes (CRISPR Associated Systems).
There are a number of slightly different mechanisms but the one that seems to be most studied and used for gene editing is cas9. My highly simplified understanding of how the defence works goes something like this:
- Hostile DNA enters the cell.
- The CRISPR/spacer sequences are transcribed into short RNA segments (CRISPR RNA or crRNA) that 'match' the spacer information and therefore (hopefully) parts of the invading DNA.
- Cas9 protein is produced which is an enzyme that can cut DNA strings.
- Cas9 binds with the crRNA fragments (along with another short bit of RNA called tracrRNA)
- The crRNA effectively guides the complex to a corresponding matching site on the invading DNA and the Cas9 cuts it at that point thus neutralising the invader!
NIce diagram of CRISPR/Cas9 - taken from the New England Biolab site
You might ask why the Cas9 complex doesn't attack the CRISPR sequences as well - which would make it a pretty useless defence mechanism. This is taken care of by the fact that the cut site DNA snippet (matched in the CRISPR spacer sequence) is chosen to be adjacent to other short sequences known as PAMs (protospacer adjacent motifs). If the Cas9 doesn't see these PAMs (which aren't present in the CRISPR/spacer sequences) then it doesn't.
Anyway, this mechanism is now being hijacked to revolutionise gene editing. In principle, the process is pretty easy:
Anyway, this mechanism is now being hijacked to revolutionise gene editing. In principle, the process is pretty easy:
- Identify the DNA sequence you want to modify.
- Build two RNA sequences that match either end of this sequence.
- Generate 2 Cas9 complexes incorporating these sequences
- Allow the mechanism to snip the faulty DNA string at the 2 sites.
- Coax the target cells repair mechanism to replace the cut-out sequence with a new, and better, sequence. There are actually a number of different ways of doing this but that's for another day.